Sequential Monte Carlo Samplers
Problem Set-Up
This is all taken from Sequential Monte Carlo samplers. We have a collection of target distributions
Importance Sampling
We write target expectations using the Importance Sampling (IS) trick for a proposal density
Sequential Importance Sampling
In importance sampling, for each different target
- At time
- Suppose that at time
In general, we cannot evaluate
In general
SMC sampler
Since the problem is integration with respect to
where we have defined the incremental weight as
To summarize:
- Importance Sampling at time
- Sequential Importance Sampling also targets
- SMC Samplers overcomes the problem of integrating over
Since the variance of the weights increases as
The algorithm is summarized below.
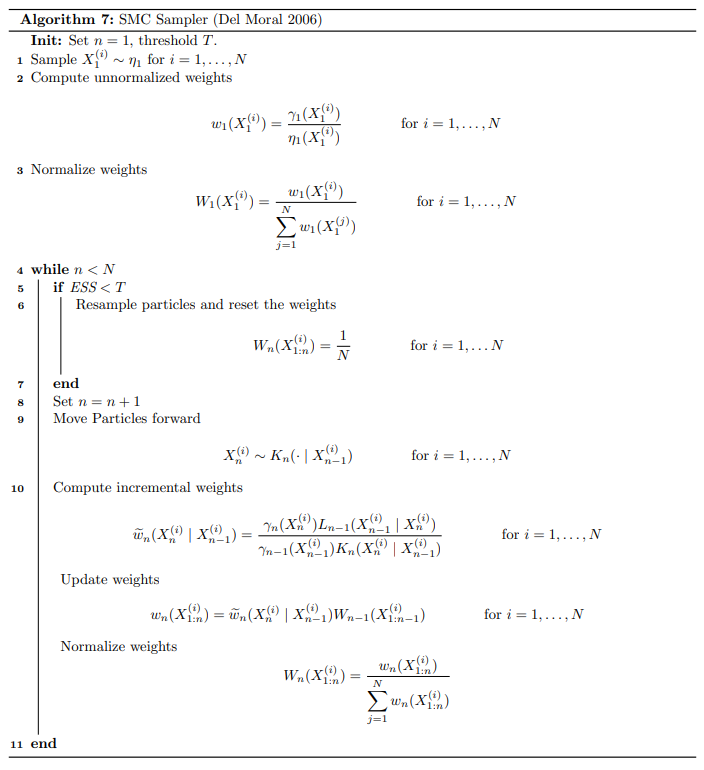
A few notes:
- The particle estimate of the
- It is helpful to remember the distributions of
- The optimal backward kernel takes us back to IS on
- Sub-optimal kernel: substitute
IS Measure Theory
Suppose
Now suppose you have samples
SIS Proposal
Now let
SMC Proposal
The proposal in the SMC sampler is not given by
SMC Steps
- Step
- Step
Define
- Step